Linjie Lu , Tristan Guyomar, Quentin Vagne, Rémi Berthoz, Alejandro Torres-Sánchez, Michèle Lieb, Cecilie Martin-Lemaitre, Kobus van Unen, Alf Honigmann, Olivier Pertz, Guillaume Salbreux, Daniel Riveline
Abstract
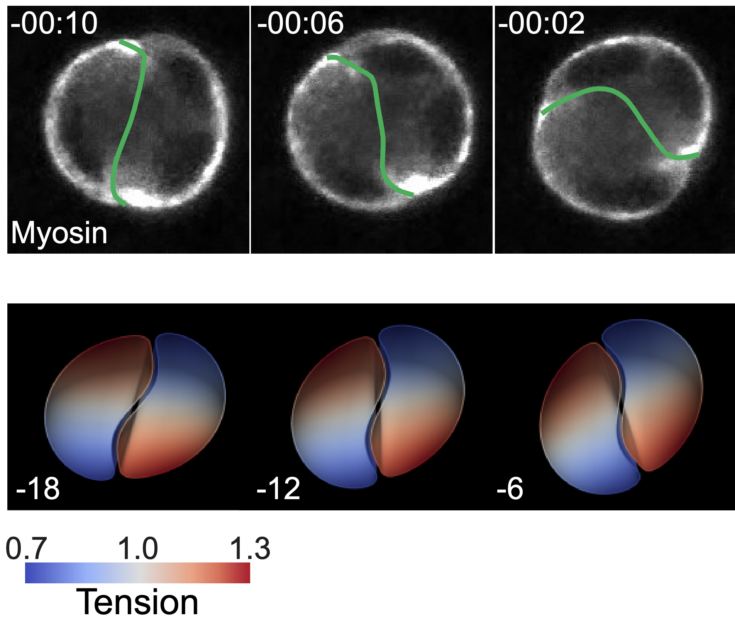
Cell mechanical interactions play a fundamental role in the self-organisation of organisms. How these interactions drive coordinated cell movement in three-dimensions remains unclear. Here we report that cell doublets embedded in a 3D extracellular matrix undergo spontaneous rotations and we investigate the rotation mechanism using live cell imaging, quantitative measurements, mechanical perturbations, and theory. We find that rotation is driven by a polarized distribution of myosin within cell cortices. The mismatched orientation of this polarized distribution breaks the doublet mirror symmetry. In addition, cells adhere at their interface through adherens junctions and with the extracellular matrix through focal contacts near myosin clusters. Using a physical theory describing the doublet as two interacting active surfaces, we find that rotation is driven by myosin-generated gradients of active tension, whose profiles are dictated by interacting cell polarity axes. We show that interface three-dimensional shapes can be understood from the Curie principle: shapes symmetries are related to broken symmetries of myosin distribution in cortices. To test for the rotation mechanism, we suppress myosin clusters using laser ablation and we generate new myosin clusters by optogenetics. Our work clarifies how polarity-oriented active mechanical forces drive collective cell motion in three dimensions.